Comparison of the NMR Spectroscopy Solution Structure of Pyranosyl-RNA and Its Nucleo-delta-peptide Analogue
For all biological systems, nature has chosen ribo- and deoxyribonucleic acids as its genetic building block. In order to understand this selectivity, the structures of the potential alternatives to the natural nucleic acids have to be investigated. We have determined the solution structures of pRNA and Nucleo-d-peptides by NMR spectroscopy. The structures pose important questions about the origin of helicity, stacking and inclination of these oligomers.
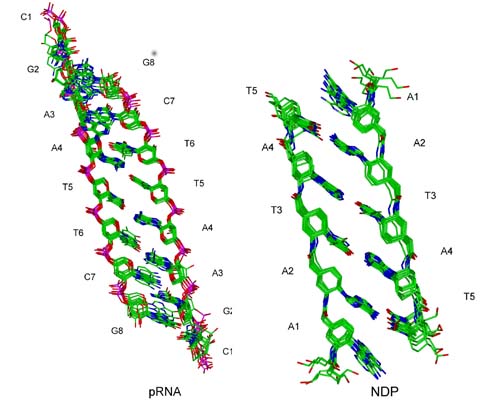
Structure of pRNA and NDP (Ilins et al., ChemBioChem 2001, in press; Schwalbe et al., Helv. Chim. Acta 83, 1079-1107 (2000)) in collaboration with groups of Prof. Quinkert (Univ. Frankfurt), Prof. Eschenmoser and Prof. Jaun (ETH Zürich).
NMR spectroscopy is used to determine the structure of proteins, of RNA and DNA in order to provide structural insight into intermolecular interactions.
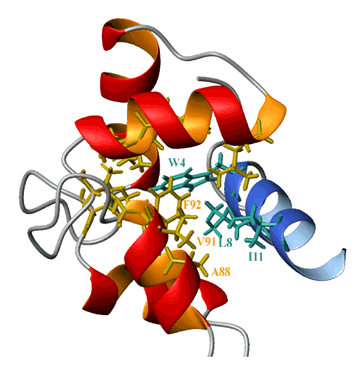
Structure of calmodulin complexed with the target peptide C20W from the Ca-ATPase pump (Elshorst et al., Biochemistry 38, 12320-12332 (1999)) in collaboration with groups of Prof. Griesinger (MPI Göttingen) and Prof. Carafoli (ETH Zürich).
